This application offers visualisation of the dynamic changes in allelic expression during early cortical development.
The neurodevelopment trajectory was built using data from day 12 mouse cortical organoids generated from F1 hybrid epiblast cells
derived from crosses between standard laboratory mice (C57BL/6J - Bl6) and four wild-derived mouse strains: Cast/EiJ, Molf/Eij,
Pwk/PhJ and Spret/EiJ.
To capture changes in allelic expression changes during cortical differentiation from radial glial cells,
pseudotemporal analyses were performed using Palantir (Setty, et al., 2019). Next, the cells were divided
into five equally-sized groups based on their pseudotime assignment, from early to the late time points (T1, T2, T3, T4 and T5).
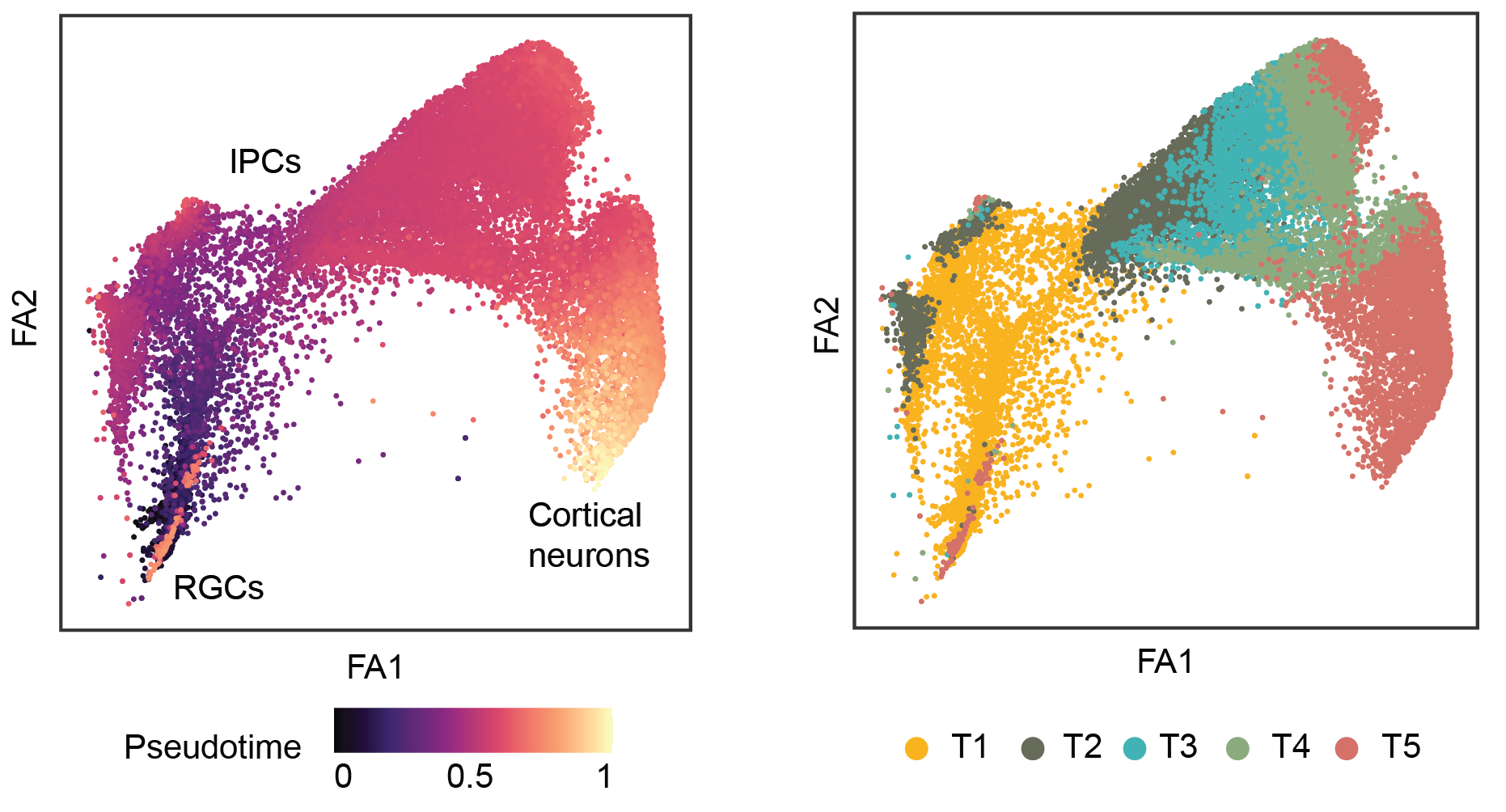
Only genes with sufficient expression (a minimum of 5 UMI counts in at least 15 cells from the neurogenic lineage: radial glial cells - RGCs,
intermediate progenitors - IPCs and cortical neurons) were included in the analysis.
A gene was considered to have differential ASE along pseudotime if the allelic distribution parameters estimated across all cells differ
from the combined estimations within each time point (significance was determined by the likelihood ratio test, FDR values provided).
Each gene is shown across four F1 mouse hybrids: Bl6Cast, Bl6Molf, Bl6Pwk and Bl6Spret. The bottom panel represents the combined
four wild-derived alleles versus the BL6 allele.
The boxplots in the top panel show the expression of each allele in the respective pseudotime group.
Differences in mean expression between the alleles were tested with a two-sided Wilcoxon test.
Summary tables contain details per clone at each time point, including mean gene expression,
allelic ratio and allelic variation (dispersion estimate) based on the pseudo-bulked counts.
To start, please select a gene of interest from the drop-down menu above.